Molecular Phylogeney and Evolutionary Relationship among Four Mosquito (Diptera: Culicidae) Species from India Using PCR-RFLP 
 Author
Author  Correspondence author
Correspondence author
Journal of Mosquito Research, 2013, Vol. 3, No. 8 doi: 10.5376/jmr.2013.03.0008
Received: 28 Feb., 2013 Accepted: 26 Mar., 2013 Published: 22 Apr., 2013
Sharma et al., 2013, Molecular Phylogeney and Evolutionary Relationship among Four Mosquito (Diptera: Culicidae) Species from India Using PCR-RFLP, Journal of Mosquito Research, Vol 3, No.8 58-64 (doi: 10.5376/jmr.2013.03.0008)
Mosquito of Culicidae (Diptera) family are known to transmit various diseases of public health importance. They are cosmopolitan in their distribution with diverse habitat and environmental conditions including in India. The generic and suprageneric relationships and the validity and monophyly of the generic and subgeneric groupings of Culicidae are in need of extensive reexamination. The application of explicit methods of phylogenetic analysis has revealed limitations in the conventional classification of mosquitoes. With the advent of molecular tools a little progress has been made, that provided a robust, stable classification that reflects evolutionary relationship. The present study is designed to establish the phylogenetic relation among the four different Culicidae mosquito species Anopheles stephensi, Aedes aegypti, Aedes albopictus and Culex quinquefasciatus by using Polymerase chain reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) of Cytochrome Oxidase I (COI) gene. The phylogenetic tree evolved using POPGENE software revealed phylogenetic relationship among them. The integration of DNA based methods foster the resolution of non-scientific but important uncertainty of nomenclature and also provide suitable means for delimiting and unambiguously identification of species for practical reasons including vector management.
1 Background
Many species of mosquitoes are vectors of pathogens that cause disease in humans and animals. The pathogens transmitted by mosquitoes include viruses (arboviruses), filaria1 worms (helminths) and protozoa. Fewer than 150 species, largely confined to genera Anopheles, Aedes (traditional broad sense) and Culex, are the indirect cause of more morbidity and mortality among humans than any other group of organisms (Severson et al., 2012; Taraphdar et al., 2012). Despite their medical importance and long history of study, the taxonomy of mosquitoes is far from complete and the existing system of classification is not entirely natural (Harbach et al., 1998; Reinert et al., 2009; Reinert et al., 2006). Major vector species are generally those most thoroughly described by conventional taxonomy. The generic and suprageneric relationships and the validity and monophyly of the generic and subgeneric groupings of Culicidae are in need of extensive reappraisal (Harbach et al., 1668). There is strong morphological and molecular evidence that subfamily Anophelinae and tribes Aedini, Culicini and Sabethini of subfamily Culicinae are monophyletic, but the other taxonomic groupings are not demonstrably monophyletic or have not been subjected to phylogenetic analyses. Mosquitoes, family Culicidae, comprise a monophyletic taxon belonging to order Diptera (Harbach et al., 1668). The family is a large and abundant group that occurs throughout temperate and tropical regions of the world, and well beyond the Arctic Circle. Some 3 490 species are currently formally recognized (Harbach et al., 2007). The integration of molecular methods could provide the support of non-scientific but important issue of classification and provides a testable means of delimiting and unambiguously identifying species for practical reasons to learn about their bionomics, distribution and relationships to disease transmission (Talbalaghl et al., 2011). This knowledge is essential for epidemiological studies, the design and implementation of appropriate vector control measures and the development of strategies for monitoring the spatial-temporal fluctuations of vector species that are needed to assess the potential risk of malaria outbreaks. In the present study the attempts have been made to generate the genetic profile of the laboratory populations of four different mosquito species Anopheles stephensi (An. Stephensi), Aedes aegypti (Ae. aegypti), Aedes albopictus (Ae. Albopictus) and Culex quinquefasciatus (Cx. quinquefasciatus) by using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of cytochrome oxidase I (COI) gene and to understand the phylogenetic relationship among them. COI gene is the mitochondrial marker often used for evolutionary study since it is the largest among the three subunits and its protein sequence contains highly conserved functional domains and variable regions. The amplified COI gene was subjected for restriction digestion by MboI, DraI, TaqI, BglII, EcoRI and MspI as single and double digestions. The results obtained from double digestions with the combination of MboI and DraI were able to differentiate among the four mosquito species. The banding patterns were analyzed using POPGENE 1.31 software and phylogenetic tree was constructed. The Aedes and Culex belong to the Culicinae family were separated from the Anopheles species and among themselves based on their phylogeny and distance.
2 Results
2.1 Genomic DNA extraction and PCR amplification of COI gene
The genomic DNA from single mosquito was isolated as described in our previous work Sharma et al. 2009 and analyzed on 0.8% agarose gel for purity. PCR amplification of COI gene led to an approximately 580 bp fragment. PCR product of the COI gene was subjected to PEG-NaCl purification for digestion purpose. The purified product represented by a single band of 580 bp with no impurities of primer-dimer, RNA etc. was further used for digestion with several restriction enzymes for the genetic analysis by PCR-RFLP. The restriction enzymes were chosen on the basis of COI gene sequences in the laboratory.
2.2 Single digestion
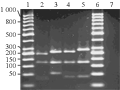
Single digestion was performed using the purified COI gene with enzymes DraI, EcoRI, TaqI, MboI, BglII and MspI (Table 1). For all digestions the fragments were visualized on 2.5% agarose gel. The restriction enzymes EcoRI and BglII were not able to digest the COI gene for any of the four species of mosquitoes under study whereas DraI produced two fragments of equal sizes with Cx. quinquefasciatus, Ae. albopictus, An. stephensi, at 280 bp & 300 bp. DraI restriction enzymes could not digest the COI gene in Ae. aegypti. TaqI digested three mosquito species with 80 bp, 200 bp & 300 bp in Cx. quinquefasciatus, 200 bp & 380 bp fragments in Ae. albopictus, 280 bp & 300 bp in An. stephensi. However no digestion was observed in Ae. aegypti. MboI was the only restriction enzyme that could digest all the COI gene of four mosquito species with digested fragments of 90 bp, 190 bp & 290 bp in Cx. quinquefasciatus, 120 bp, 180 and 280 bp in both Ae. aegypti and Ae. albopictus while in An. stephensi fragments were of 190 bp, 390 bp. MspI digested the gene into two fragments of 180 bp, 400 bp in Ae. aegypti and 260 bp & 320 bp in Ae. albopictus. But no digestion was observed in Cx. quinquefasciatus and An. stephensi. The respective fragment sizes with all the restriction enzymes as single digestion have been summarized in Table 1.
![]()
Table 1 Single digested fragments of COI gene of Cx. quinquefasciatus, Ae. aegypti, Ae. albopictus, An. stephensi with DraI, TaqI, MboI, MspI, BglII and EcoRI
2.3 Double digestion
When double digestion was performed using different combinations of MboI+ DraI, and MboI + TaqI. The double digestion with MboI+TaqI revealed that Ae. aegypti, Ae. albopictus, An. stephensi are similar in their restriction pattern with visible fragments of 120, 180 & 280 bp sizes whereas a replacement of 90 bp band in place of 120 bp in Cx. quinquefasciatus which differentiated it from all other mosquito species under study. The combinations of MboI+ DraI used were noteworthy as the digestion with these restriction endonucleases was capable of differentiating all four mosquito species under study from each other. The double digested fragments of all the four species have been summarized in Table 2.
.png) Table 2 Double digested products of COI gene of Cx. quinquefasciatus, Ae. aegypti, Ae. albopictus and An. stephensi with a combination of MboI+ DraI and MboI+ TaqI enzymes |
2.4 Estimation of polymorphism and construction of phylogenetic tree
Based on the double digestion with MboI+ DraI the genetic polymorphism among the various mosquito species was evaluated using the POPGENE 1.31 version. A total of 8 bands or loci representing all the four mosquito species were considered (Figure 1). The percent polymorphism obtained was 87.5% among the different mosquito species. The genetic diversity among the species indicated by the Shannon Index and the gene diversity index of 0.34 each as shown in Table 3. The minimum genetic distance was obtained between Cx. quinquefasciatus and Ae. albopictus of 0.28, whereas the maximum genetic identity was also observed as 0.75 between these two species (Table 4). Consensus tree generated by using the data showed that Aedes species (Ae. aegypti and Ae. albopictus) and Cx. quinquefasciatus are closely placed (Figure 2). On the other hand An. stephensi is represented in the separate branch with maximum distance of 0.69 with the other three mosquito species and having least identity.
|
Table 4 Nei's Original Measures of Genetic Identity and Genetic distance based on MboI+ DraI double digestion |
3. Discussion
The first evolutionary tree of Culicidae was constructed by Ross (Ross, 1951) based on an intuitive interpretation of comparative bionomics and morphological data. The tree reflected the traditional division of family Culicidae into three subfamilies: Anophelinae, the basal lineage; Toxorhynchitinae, the intermediate lineage; and Culicinae, the large derived lineage. The relationships illustrated by Ross (Ross, 1951) are those more or less traditionally accepted and unchallenged by later workers until Reinert, Harbach and Kitching (Reinert et al., 2009) examined the generic relationships and higher classification of the family based on cladistic analyses of morphological data. Reidenbach et al., 2009 examined the phylogenetic relationships of four mosquito species, one from each of genera Aedes, Anopheles, Culex and Toxorhynchites, using 18S and 5.8S rDNA sequences. Their analysis placed Anopheles in a position basal to the other species. In a parsimony analysis- Toxorhynchites was placed as the sister group of Aedes + Culex, whereas Aedes was the sister group of Toxorhynchites + Culex in a maximum likelihood tree.
However, neither position was strongly supported by bootstrapping. The use of neighbour-joining procedures produced a trichotomy for the three species. Consequently, the results of these taxon-limited analyses neither support nor refute the traditional position of Toxorhynchites as the sister group of subfamily Culicinae (Reidenbach et al., 2009). Considering the impact of mosquitoes on human health, it is not surprising that phylogenetic studies of these insects have only dealt with relationships within the four major groups that contain the majority of medically important species. These include subfamily Anophelinae and tribes Aedini, Culicini and Sabethini of subfamily Culicinae. Mosquitoes, because of their medical importance, are one of the most thoroughly studied groups of insects. Ironically, despite the significant amount of morphological and molecular work that has been done, little progress has been made toward an understanding of the evolutionary history of the family. As the synthesis of available information clearly shows, the absence of a comprehensive phylogeny for mosquitoes is a major obstacle to realizing a robust, stable classification of the family.
PCR-RFLP is an efficient and convenient typing technique which is able to detect variation in the DNA sequence by breaking the DNA in fragments with restriction enzymes and analyzing size of resulting fragments by gel electrophoresis. PCR-RFLP has been used in the study of genetic structure of malaria vector Anopheles subpictus in Iran using mitochondrial cytochrome oxidase COI and COII (Karimian et al., 2011). Similar technique has been used to distinguish between several Diabrotica species using PCR-RFLP of the COI gene in which mitochondrial COI subunit was amplified and digested (Alam et al., 2012). The comparative linkage map for Culex pipiens (Cx. pipiens) and Ae. aegypti was also developed based on RFLP using cDNA clones of Ae. aegypti as probes to southern blot of Cx. pipiens genomic DNA.
Present study reveals that PCR-RFLP is a versatile molecular tool to screen genetic polymorphism among various mosquito species. The PCR-RFLP has been able to demonstrate clear genetic diversity by using the COI gene in the present study. The use of this gene is arbitrary and it is highly probable that other mitochondrial protein coding genes can be used to differentiate these species. In preliminary studies using other mitochondrial genes such as 16S rRNA, COII and ND5 it was observed that many of the mosquito species could be differentiated in a similar manner (Alam et al. 2012; Ozdil et al., 2012; Zitco et al., 2011).
Double digestion performed with combination of enzymes MboI+ DraI during the PCR-RFLP provided significant genetic variation among the four mosquito species under study reveals that Aedes species (Ae. aegypti, Ae. albopictus) are closely related to the Cx. quinquefasciatus as minimum genetic distance and maximum genetic identity was observed among them. The finding can be further substantiated by the facts that these three belongs to the same family Culicidae. Moreover, at the structural and behaviour level also they share many characters such as their sitting posture, larval movement in water, position of siphon etc. Amongst the two Aedes sps., the Ae. albopictus is found more closer to Cx. quinquefasciatus compared to Ae. aegypti, having a genetic distance of 0.28. It may be well thought-out that they might have evolved from a similar origin during evolutionary process. The study undertaken proposes that Ae. aegypti and Ae. albopictus has evolved in the same line but Ae. albopictus has diversed into another independent line as the Cx. quinquefasciatus species. The most genetically diverse species, the An. stephensi has shown maximum genetic distance to other three species. Thus it can be considered as the most distantly related species among all species under study. Whether the outcomes of our study are factual and can be established with extensive assessment of literature and studies carrying out by using some other molecular markers. The insights of the genetic pattern of a vector species are fundamental in understanding the diseases epidemiology. Such an understanding becomes crucial in devising vector control strategies.
4. Materials and Methods
4.1. Test insect
Laboratory reared Cx. quinquefasciatus, Ae. aegypti, Ae. albopictus, An. stephensi were used as test insect. The adult mosquitoes were collected with the help of mechanical aspirator from the stock colony maintained in the laboratory at (27± 2)℃ and relative humidity of (70 ± 5)%.
4.2. Extraction of mosquito DNA
The DNA extraction was done by using the method as described in our previous publication (Sharma et al., 2009). In this each single adult mosquito was homogenized in the micro centrifuge tube by adding 100 µL lysis buffer. The homogenate was immediately kept on ice for 10 minutes and followed by heat treatment at 65℃ for 30 minutes. Subsequently, 30 µL 5 M potassium acetate was added and immediately transferred to ice for one hour followed by centrifugation at 13 000 rpm for 15 minutes at 10℃. To the supernatant obtained, a double volume of absolute chilled ethanol was added for precipitation of DNA and kept at -20℃ for overnight. After centrifugation at 13 000 rpm for 15 minutes at 10℃, the precipitated DNA was washed in 70% ethanol twice. The palleted DNA was allowed to air dry and finally dissolved in 50 µL TE buffer.
4.3. PCR amplification of COI gene
The PCR amplification reactions were performed in 25 µL reaction mixture per tube containing 2.5 µL of 10× assay buffer, 2.5 mM of MgCl2, 2 mM each dNTP, 10 µL of forward and reverse primers for COI gene and 10 ng of genomic DNA. Reactions were performed in a (BIORAD PCR System iCycler) thermal cycler. The PCR condition consisted of initial denaturation step for 4 min at 94℃ followed by 35 cycles of 1 min at 94℃, 1 min at 40℃, and 2 min at 72℃. A final extension step was performed at 72℃ for 10 min. A negative control (PCR mix without DNA) was also prepared alongwith the other samples.
4.4. Purification of PCR-product
The 0.6 volume of PEG-NaCl was added into the PCR-product. After a brief vortexing the samples were incubated at 37℃ for 30~40 minutes followed by centrifugation at 10 000 rpm for 30 minutes. The pellet was washed twice by adding 100 µL of 70% chilled ethanol followed by centrifugation at 10 000 rpm for 30 minutes. Finally, the pellet was allowed to air dry and re-suspended in 10 µL of triple distilled water.
4.5. Restriction digestion of PCR product
4.5.1. Single digestion
The purified PCR product of four species were digested using 10 µL of nuclease free water, 2 µL of 10 X fast digest buffer, 2 µL of DNA, and 1 µL of fast digest enzymes DraI, EcoRI, TaqI, MboI and BglII at 37℃ for 5 minutes. A volume of 15 µL of control was also prepared with all the above components. Followed by incubation at 65℃ was carried out for 5 minutes to inactivate the enzyme.
4.5.2. Double digestion
Double digestion was performed in a reaction volume of 18 µL consisting 10 µL of nuclease free water, 2 µL of 10 X Fast Digest Buffer, 2 µL of DNA sample and 2 µL of each enzyme was added. A combination of MboI + DraI and MboI + TaqI was taken. Double digestion was performed by the procedure similar to that of single digestion.
4.6. Statistical analysis
Polymorphism in above mosquito species was analyzed by scoring the polymorphic and monomorphic bands. If same band of DNA is present in all indivisuals or the sample populations under study it is said to be monomorphism whereas in polymorphism is defined as discontinuous variation in a single population. The genetic distance and the polymorphism among the population were interpreted by using POPGENE version 1.31 software. A suitable dendogram was generated with the help of above software.
Authors' Contributions
Sharma A.K.: The main author of the paper and have made contribution to conception and design and also the analysis of data; Tyagi V: This author is involved in the acquisition of data by using various methods; Chandel K: Helped in the general supervision of the research group; Mendki MJ: Involved in drafting of Manuscript; Tikar SN: Revising the MS critically for important intellectual contents; Sukumaran D: Have given final approval of the version to be published.
Acknowledgement
The authors are thankful to Prof. (Dr.) M. P. Kaushik, Director, Defence Research & Development Establishment, Gwalior, Madhya Pradesh, India for providing all necessary facility to conduct this research work. Sincere thanks also to the scientists and supportive staff of Vector Management Division for their kind cooperation for carrying out the above work.
References
Alam M.S., Kato H., Fukushige M., Wagatsuma Y., and Itoh M., 2012, Application of RFLP-PCR- based identification for sand fly surveillance in an area endemic for kala azar in mymensingh, Bangladesh, J. Parasitol Res., article id 467821
http://dx.doi.org/10.1155/2012/467821 PMid:22701164 PMCid:3369511
Harbach R.E., and Kitching I.J., 1998, Phylogeny and classification of the Culicidae (Diptera), Syst. Entomol., 23: 327-370
http://dx.doi.org/10.1046/j.1365-3113.1998.00072.x
Harbach R., 2007, The Culicidae (Diptera): A review of taxonomy, classification and phylogeny, Zootaxa, 1668: 591-638
Harbach R.E., and Howard T.M., 2007, Index of currently recognized mosquito species (Diptera: Culicidae), Eur. Mosquito Bulletin, 23: 1-66
Karimian F., Sedaghat M.M., Oshaghi M.A., Mohtarami F., Dehkordi A.S., Koosha M., Akbari S., and Hashemi- Aghdam S.S., 2011, Utility of filter paper for preserving insects, bacteria, and host reservoir DNA for molecular testing, Iran J. Arthropod-Borne Dis., 5(2): 42-50
PMid:22808417 PMCid:3385577
Ozdil F., Aytekin I., Ilhan F., and Boztepe, 2012, Genetic variation in Turkish honeybees Apis mellifera anatoliaca, A. m. caucasica, A. m. meda (Hymenoptera: Apidae) inferred from RFLP analysis of three mtDNA regions (16S rDNA-COI-ND5), Eur. J. Entomol., 109(2): 161-167
Reidenbach K.R., Cook S., Bertone M.A., Harbach R.E., Wiegmann B.M., and Besansky N.J., 2009, Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology, BMC Evol. Biol., 298(9): 1-14
Reinert J.F., Harbach R.E., and Kitching I.J., 2006, Phylogeny and classification of Finlaya and allied taxa (Diptera: Culicidae: Aedini) based on morphological data from all life stages, Zoological Journal of the Linnean Soc., 148: 1-100
http://dx.doi.org/10.1111/j.1096-3642.2006.00254.x
Reinert J.F., Harbach R.E., and Kitching I.J., 2009, Phylogeny and classification of tribe Aedini (Diptera: Culicidae), Zoo. J. Linnean Soc., 157: 700-794
http://dx.doi.org/10.1111/j.1096-3642.2009.00570.x
Severson D.W., and Behura S.K., 2012, Mosquito genomics: progress and challenges. Annu. Rev. Entomol., 57: 143-166
http://dx.doi.org/10.1146/annurev-ento-120710-100651 PMid:21942845
Sharma A.K., Mendki M.J., Tikar S.N., Chandel K., Sukumaran D., Parashar B.D., Vijay Veer, Agarwal O.P., and Prakash S., 2009, Genetic variability in geographical populations of Culex quinquefasciatus Say (Diptera: Culicidae) from India based on random amplified polymorphic DNA analysis, Acta Tropica, 112: 71-76
http://dx.doi.org/10.1016/j.actatropica.2009.06.014 PMid:19577531
Ross H.H., 1951, Conflict with Culex, Mosquito News, 11: 128-132
Talbalaghl A., and Shaikevich E., 2011, Molecular approach for identification of mosquito species (Diptera; Culicidae) in province of alessandria, piedmont, Italy, Eur J Entomol., 108(1): 35-40
Taraphdar D., Sarkar A., and Chatterjee S., 2012, Mass scale screening of common arboviral infections by an affordable, cost effective RT-PCR method, Asian Pac. J. Trop. Biomed., 2: 97-101
http://dx.doi.org/10.1016/S2221-1691(11)60200-1
Zitko T., Kovacic A., Desdevises Y., and Puizina J., 2011, Genetic variation in East-Adriatic populations of the Asian tiger mosquito, Aedes albopictus (Diptera: Culicidae), inferred from NADH5 and COI sequence variability, Eur. J. Entomol., 108(4): 501-508
. PDF(130KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Ajay Kumar Sharma
. Chandel K.
. Tyagi V.
. Mendki M.J.
. Tikar S.N.
. Sukumaran D.
Related articles
. Culicidae
. Mosquitoes
. PCR-RFLP
. Cytochrome oxidase-I
Tools
. Email to a friend
. Post a comment
.png)